Development of a new class of liver receptor homolog-1 (LRH-1) agonists by photoredox conjugate addition.
Cornelison, J.L., Cato, M.L., Johnson, A.M., D'Agostino, E.H., Melchers, D., Patel, A.B., Mays, S.G., Houtman, R., Ortlund, E.A., Jui, N.T.(2020) Bioorg Med Chem Lett 30: 127293-127293
- PubMed: 32631515
- DOI: https://doi.org/10.1016/j.bmcl.2020.127293
- Primary Citation of Related Structures:
6VIF - PubMed Abstract:
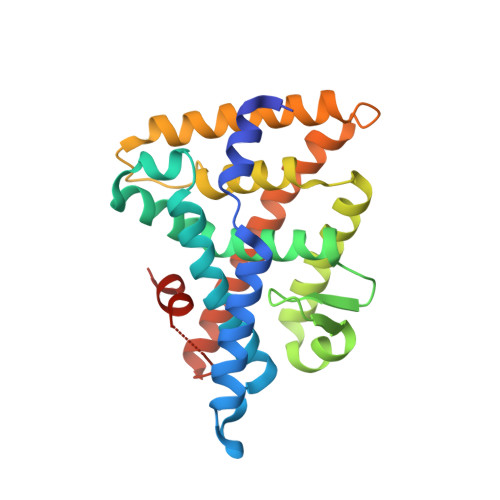
LRH-1 is a nuclear receptor that regulates lipid metabolism and homeostasis, making it an attractive target for the treatment of diabetes and non-alcoholic fatty liver disease. Building on recent structural information about ligand binding from our labs, we have designed a series of new LRH-1 agonists that further engage LRH-1 through added polar interactions. While the current synthetic approach to this scaffold has, in large part, allowed for decoration of the agonist core, significant variation of the bridgehead substituent is mechanistically precluded. We have developed a new synthetic approach to overcome this limitation, identified that bridgehead substitution is necessary for LRH-1 activation, and described an alternative class of bridgehead substituents for effective LRH-1 agonist development. We determined the crystal structure of LRH-1 bound to a bridgehead-modified compound, revealing a promising opportunity to target novel regions of the ligand binding pocket to alter LRH-1 target gene expression.
Organizational Affiliation:
Department of Chemistry, Emory University, Atlanta, GA 30322, United States.